Spreader events and the limitations of projected networks for capturing dynamics on multi-partite networks
Abstract
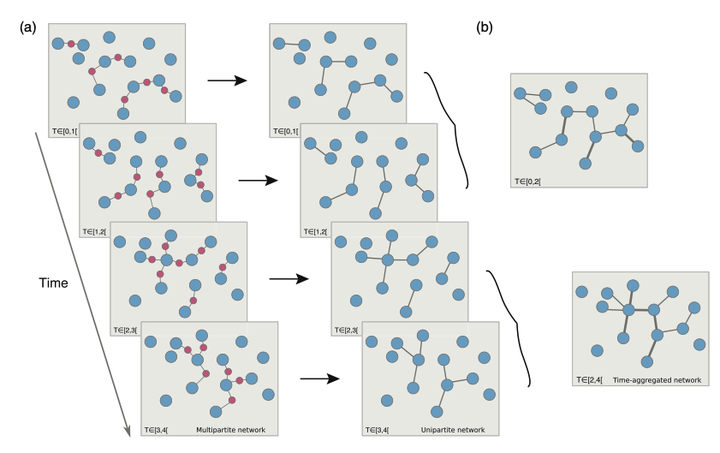
Many systems of scientific interest can be conceptualized as multipartite networks. Examples include the spread of sexually transmitted infections, scientific collaborations, human friendships, product recommendation systems, and metabolic networks. In practice, these systems are often studied after projection onto a single class of nodes, losing crucial information. Here, we address a significant knowledge gap by comparing transmission dynamics on temporal multipartite networks and on their time-aggregated unipartite projections to determine the impact of the lost information on our ability to predict the systems' dynamics. We show that the dynamics of transmission models can be dramatically dissimilar on multipartite networks and on their projections at three levels: final outcome, the magnitude of the variability from realization to realization, and overall shape of the temporal trajectory. We find that the ratio of the number of nodes to the number of active edges over the time aggregation scale determines the ability of projected networks to capture the dynamics on the multipartite network. Finally, we explore which properties of a multipartite network are crucial in generating synthetic networks that better reproduce the dynamical behavior observed in real multipartite networks.